Part 2: GeoFLOW - Automate GeoSpatial workflow using Notebook & Papermill
Executing notebooks end-to-end & keeping a separate copy for each instance is a tedious & ugly task. Can we automate this & make our workflow better?
Problems with Notebook
In part 1 of the GeoFLOW series, we created a GeoSpatial workflow using Python & Jupyter Notebook. Looking at the intermediary visuals is certainly useful but there are a few drawbacks in creating such workflows:
- Version Controlling Notebooks are difficult to version control, we can eliminate a part of the pain using nbdime but still, you will add up to your commits every time you open a notebook or click on a cell.
- Scalability For every city you wish to repeat the workflow, you have to manually run the cells one at a time. The next alternative is to move everything to a python script & save the images to inspect later. (I don’t like either)
Can we do any better?
Papermill gives us the superpowers to parameterize & execute the notebooks.
How does this solve our problems:
- Version Controlling We always clean the outputs of the parameterized notebook before committing the code. This keeps our workflow & commit messages sane.
- Scalability Now we can execute notebooks by running a python scripts, it will have our notebooks ready & baked for further visual analysis.
This workflow also makes it easier for us to catch bugs & fix them in place. (Try them out yourself, don’t just take my word for it)
How to use this technique?
We will repeat the same process that we had defined in Part 1 of this series but now we will repeat it for multiple cities, i.e Fortportal & Entebbe.
Steps
-
Parameterize the required cell in the notebook
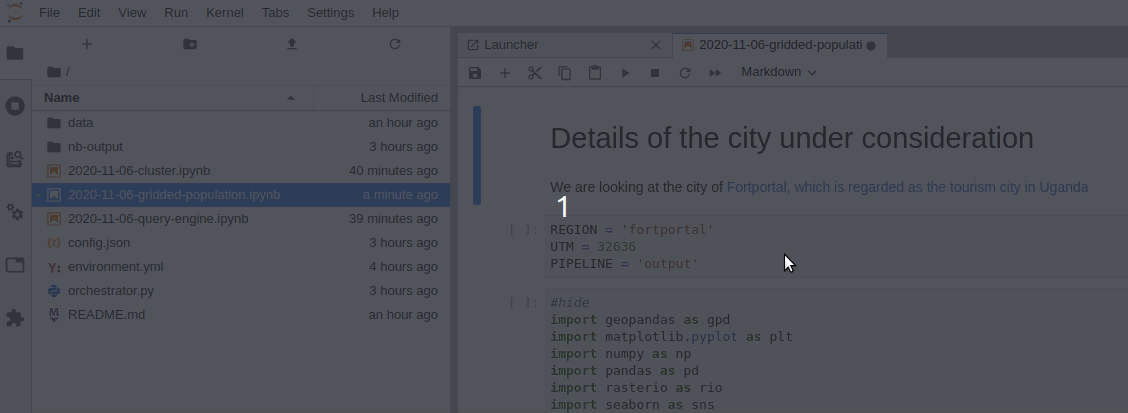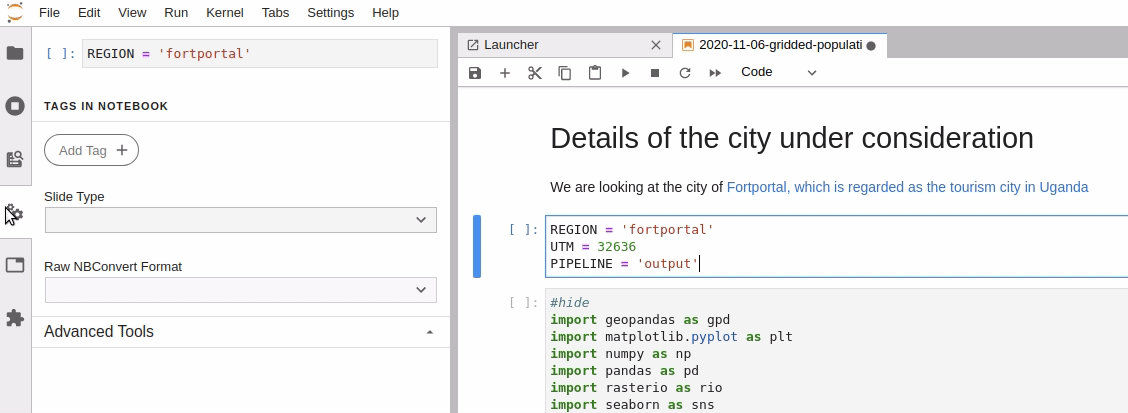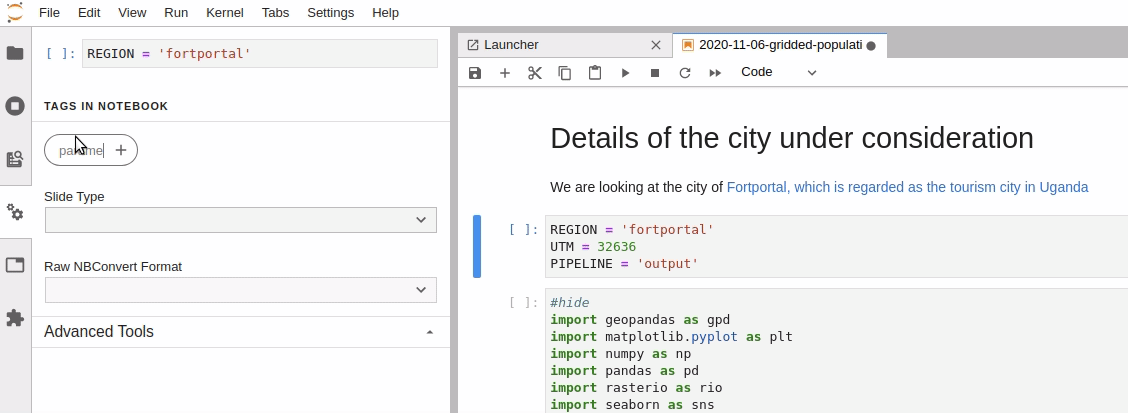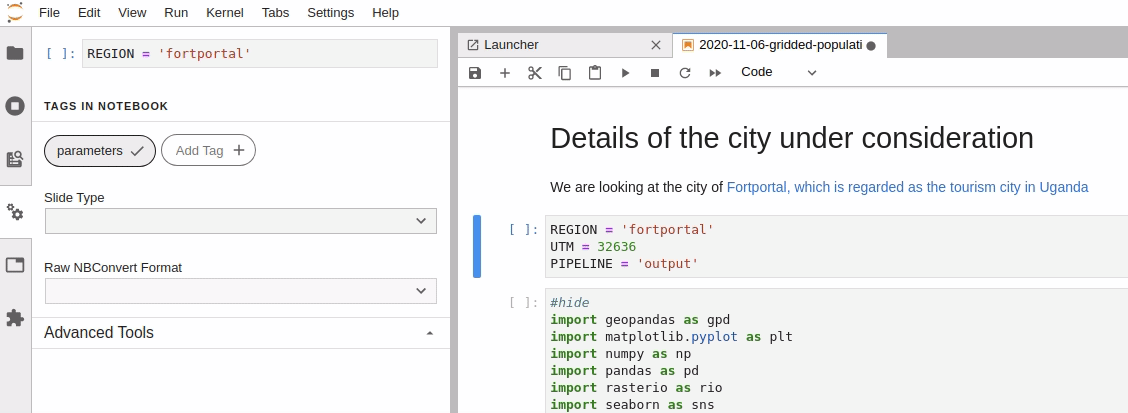
-
Define a configuration file
config.jsonthat would replace theparameterscell (i.e the cell we tagged in step 1) with the values with which we want to execute the notebook.[ { "REGION": "fortportal", "UTM": 32636, "PIPELINE": "gridded-population" }, { "REGION": "entebbe", "UTM": 32636, "PIPELINE": "gridded-population" } ] -
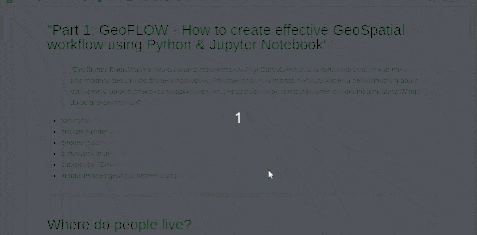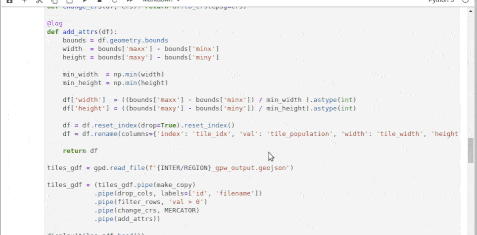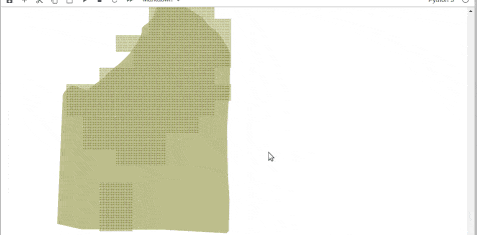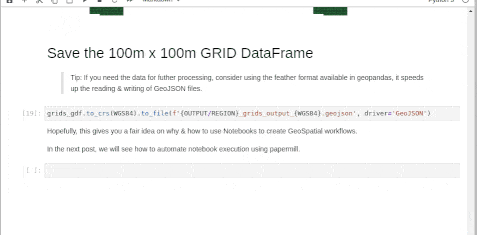
Add an
orchestrator.pyto orchestrate the workflowdef process(region, pipeline): '''Processes the pipeline for a given region & stores the executed notebook inside nb-output/ folder. Input - region: Name of the region - pipeline: Pipeline to run (gridded-population / cluster / query-engine) Return None ''' def execute(nb, pipeline, region): '''Execute a notebook using papermill.''' pm.execute_notebook( input_path=nb, output_path=f'nb-output/{pipeline}-executed-{region["REGION"]}_@_{dt.now().strftime("%Y-%m-%d_%I-%M-%S_%p")}.ipynb', parameters=region ) if pipeline == 'query' : execute('2020-11-06-query-engine.ipynb' , pipeline, region) elif pipeline == 'cluster' : execute('2020-11-06-cluster.ipynb' , pipeline, region) elif pipeline == 'gridded-population': execute('2020-11-06-gridded-population.ipynb', pipeline, region) else: print('Not a valid pipeline') -
Run the
orchestrator.pyfilepython orchestrator.py
This will execute the parameterized notebooks & store the executed notebooks inside nb-output/ folder.
Fortportal

Entebbe
Try this workflow & suggest any changes to improve it further. Happy Coding!